GIFT Shuangjia Zheng's Team Publishes in Nature Machine Intelligence: Geometric Deep Learning Tackles Challenge of RNA-Targeted Drug Discovery
RNA plays a crucial role in cellular regulation, and RNA-targeted small-molecule drugs are promising for expanding the traditionally protein-centric landscape of drug discovery. However, accurately modeling ligand recognition remains challenging due to RNA's highly flexible structures and complex binding modes, with existing methods facing significant limitations in predicting binding specificity and supporting drug screening.
On December 12, 2025, a research team led by Assistant Professor Shuangjia Zheng at the Global Institute of Future Technology, SJTU, published a paper titled "Deciphering RNA–ligand binding specificity with GerNA-Bind" in Nature Machine Intelligence, in collaboration with Guangdong University of Technology and Harvard University. The paper introduced GerNA-Bind, a geometric deep learning framework that integrates multistate RNA structural information with uncertainty evaluation, providing both accuracy and biological insight for the development of RNA-targeted small-molecule drugs.

RNA is integral to cellular regulation - its interactions with small molecules and proteins profoundly influence gene expression and cellular function. In recent years, RNA-targeted small-molecule drugs have been recognized as transformative strategies to broaden the scope of traditionally protein-focused drug research. Nonetheless, existing models still face limitations in predicting binding specificity and aiding drug screening due to the structural complexity of RNA. While general structural models such as AlphaFold3 are beginning to show promise, they often fall short in accurately predicting binding strength and elucidating interaction mechanisms.
To address this challenge, the research team developed GerNA-Bind, a geometric deep learning framework for predicting RNA-small molecule binding specificity. This framework integrates multistate RNA structural information with uncertainty modeling to deliver reliable predictions of RNA-ligand interactions. In a demonstration, GerNA-Bind was applied to perform conformation-specific virtual screening against the oncogenic MALAT1 RNA, successfully identifying and validating several binding micro molecules. Some of these molecules exhibited specific recognition for the triple-helix conformation, highlighting the method's potential for RNA-targeted drug discovery.
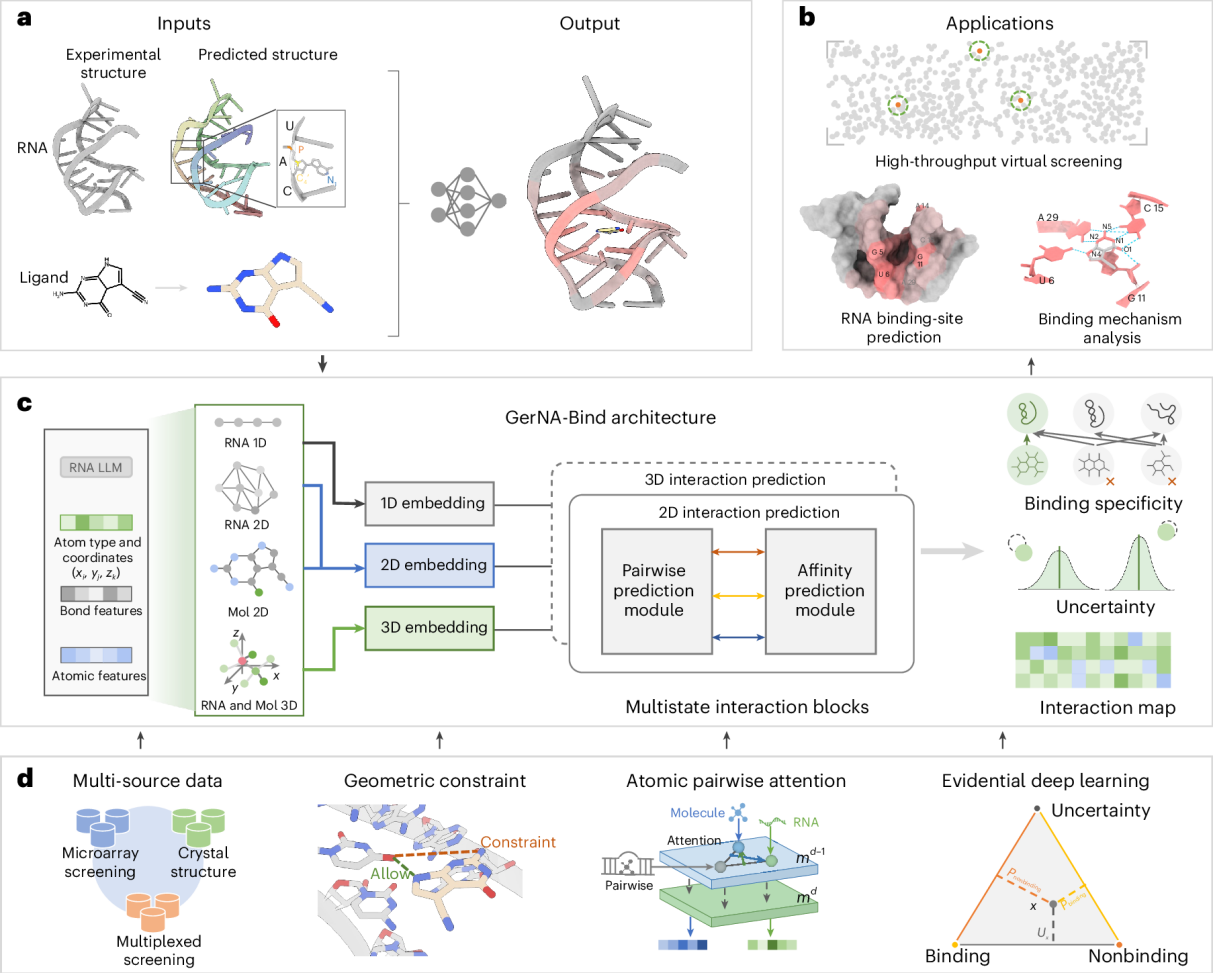
Figure 1 - Overview of GerNA-Bind
Tests on multiple benchmark datasets demonstrated GerNA-Bind's exceptional capabilities. It maintained stable and high-accuracy predictions under conditions of low homology, high sequence diversity, structural perturbations, and varying ligand complexity. This robustness stems from its unique geometric deep learning design-its geometric constraint module directly generates base-atom contact matrices, enabling the reconstruction of spatial contact patterns between RNA and small molecules without requiring external complex structures. Furthermore, GerNA-Bind provides uncertainty evaluation during prediction, offering a scientific foundation for prioritizing the most reliable molecules in virtual screening. The researchers note that this interpretable, quantifiable approach not only reveals fine-grained mechanisms of RNA-ligand interactions but also enhances the model's generalization to other RNAs.

Figure 2 - GerNA-Bind validation for RNA binding-site identification
The advantage of GerNA-Bind was further validated in a real-world drug discovery pipeline. Targeting the triple-helix conformation of the oncogenic MALAT1 RNA, the research team performed virtual screening on 21,659 candidate molecules. Using GerNA-Bind's prediction scores and uncertainty estimates, they prioritized 28 candidates, 18 of which were confirmed to bind RNA in vitro experiments. Notably, some molecules, despite their high structural novelty, showed high selectivity for MALAT1 and inhibited cancer cell migration in cellular assays, demonstrating GerNA-Bind's practical utility in discovering RNA-targeting small molecules.

Figure 3 - GerNA-Bind assists wet-lab experiments for RNA-targeting drug discovery
Yunpeng Xia, a doctoral student at Shanghai Jiao Tong University; Jiayi Li, a graduate student at Harvard University and research assistant of Zheng Lab; Yiting Chu, a graduate student at Guangdong University of Technology, are co-first authors of the paper. Professor Shuangjia Zheng is the corresponding author.
Paper Title:
Xia, Y., Li, J., Chu, YT. et al. Deciphering RNA–ligand binding specificity with GerNA-Bind. Nat Mach Intell (2025)
Paper Link:
https://www.nature.com/articles/s42256-025-01154-z
Author Profile

Yunpeng Xia
Ph.D. student at Shanghai Jiao Tong University (Class of 2025). Research interests: AI-assisted drug discovery, protein design, and cryo-EM structure determination.

Jiayi Li
Graduate student in the Department of Biostatistics at Harvard University. Research interests: AI-assisted drug discovery, multi-omics data integration, and single-cell analysis.

Shuangjia Zheng
Tenure-Track Assistant Professor and P.h.D. Supervisor of the Global Institute of Future Technology, SJTU. Prof. Zheng's research primarily focuses on the intersection of generative artificial intelligence and drug design. He has published over 50 papers in prestigious international journals and conferences, including Nat. Mach. Intell., Nat. Comput. Sci., Nat. Commun., Nat. Biomed. Eng, NeurIPS, and ICLR, with citations exceeding 5000. Several of his achievements have been reported by renowned media outlets, including MIT Tech Review, Forbes, China Science Daily, People's Daily, and Xinhua Net. He has been selected for the Asian Young Scientist Fellowship, Forbes 30 Under 30 Asia, the WAIC Yunfan Award, and the Shanghai Chenguang Program. He has also received honors and awards, including the World Artificial Intelligence Conference Outstanding Paper Award, the Rey Wu Prize, the Baidu Scholarship, and the CAAI Outstanding Doctoral Dissertation Award.




