GIFT Shuangjia Zheng team's AI RNA design research published at top AI conference NeurIPS 2025
A team led by Assistant Professor Shuangjia Zheng at Global Institute of Future Technology, SJTU has published a research paper titled "RiboFlow: Conditional De Novo RNA Co-Design via Synergistic Flow Matching" at the 39th Annual Conference on Neural Information Processing Systems (NeurIPS 2025), a top-tier academic conference on deep learning and machine learning. The co-first authors of the paper are Runze Ma (2025 Ph.D. candidate) and Zhongyue Zhang (2024 Ph.D. candidate) from GIFT. The team had a total of three papers accepted at NeurIPS this year, covering research directions such as RNA design, protein design and antibody design.
The Conference on Neural Information Processing Systems (NeurIPS) is a top-level international conference in machine learning and computational neuroscience. Ranked as one of the Category A conferences in artificial intelligence by the China Computer Federation, NeurIPS covers various sub-fields including deep learning, computer vision, large-scale machine learning, learning theory, optimization and sparse theory.
Paper link: https://arxiv.org/abs/2503.17007
Research Background
Ribonucleic acid (RNA) is an essential biological macromolecule in life sciences. It acts like a highly customized "molecular key" that can fold into specific 3D shapes to precisely bind with target molecules (e.g., drug molecules), thus playing important roles in disease treatment and biological detection. Therefore, it is a significant challenge in biomedicine to design novel RNAs that can bind with specific targets.
Current Research
Due to the high flexibility of RNA structure, the design of RNA faces several challenges: First, both the rationality of the 3D shape and the corresponding chemical sequence must be considered. Existing methods often struggle to optimize both simultaneously, resulting in sequences that mismatches the ideal structure. At the data level, high-quality data on RNA-small molecule interactions are rare, leaving models with insufficient training materials. Moreover, current methods typically fail to guide the RNA generation process due to ineffective utilization of the shape information of target molecules, which limits the ability to generate RNAs tailored to specific molecules.
Research Results
To address these challenges, this paper proposes an innovative model named “RiboFlow” that introduces a Synergistic Flow Matching algorithm. It simultaneously model the 3D structure and sequence of RNA, ensuring high compatibility between the two, and thereby designing RNA molecules with high affinity for target molecules. Additionally, the research team compiled and published RiboBind - the largest currently available dataset for RNA-small molecule interactions - effectively alleviating data shortage issues. Experimental results show that RNAs designed by RiboFlow significantly outperform existing methods in both structural rationality and target binding capability, providing an efficient and reliable framework for the development of RNA drugs and biological tools.
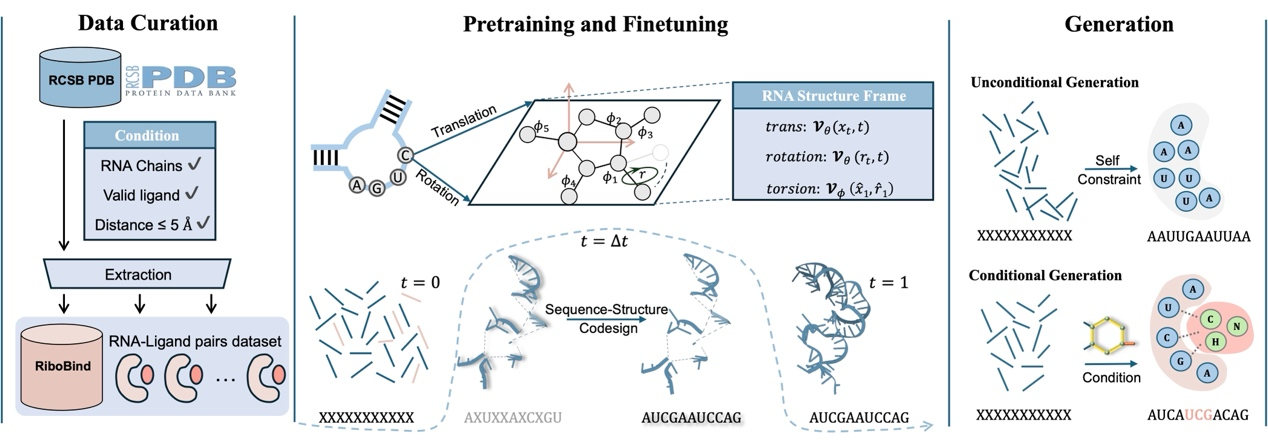
Figure 1: The authors first collected and constructed a large-scale RNA-ligand interaction dataset, RiboBind, from the RCSB PDB database. Then, they pre-trained RiboFlow on the RNAsolo dataset to enhance its geometric perception capabilities. After fine tuning, RiboFlow can design high-affinity RNAs under ligand constraints, and generate structurally valid and high-affinity RNAs for different ligands.
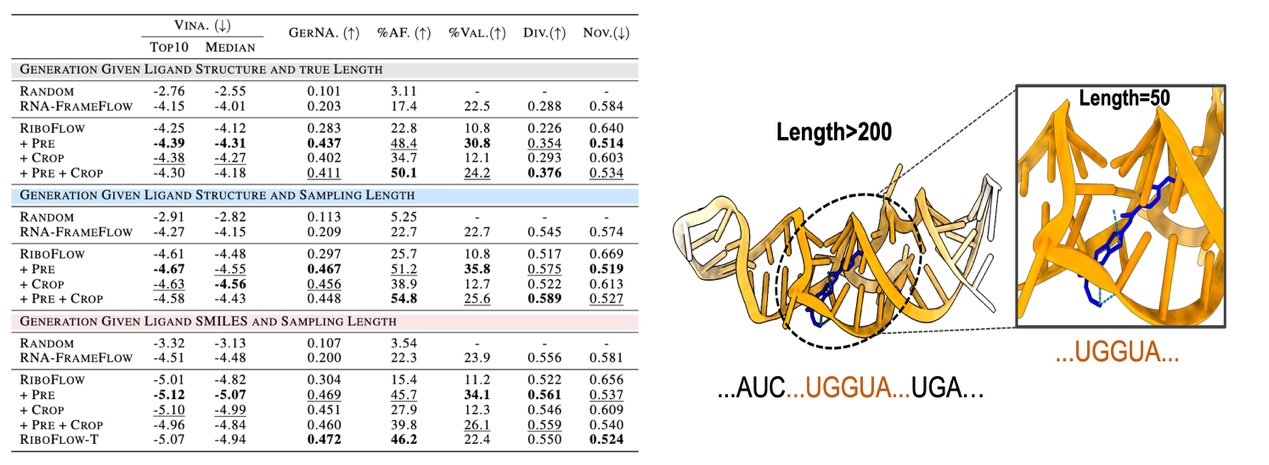
Figure 2: (Left) RiboFlow and its variants achieve optimal RNA design performance across multiple tasks. (Right) RiboFlow enables de novo design for ligands with binding capability, validating its design proficiency.
Author Profile

Runze Ma
2025 Ph.D. candidate of Global Institute of Future Technology, SJTU. Research interests: Generative artificial intelligence, AI for science.

Zhongyue Zhang
2024 Ph.D. candidate of Global Institute of Future Technology, SJTU. Research interests: Multi-agent systems, large language model applications, and generative artificial intelligence.

Shuangjia Zheng
Tenure-Track Assistant Professor and PhD supervisor of Global Institute of Future Technology, SJTU. Prof. Zheng's research primarily focuses on the intersection of generative artificial intelligence and drug design. He has published over 50 papers in prestigious international journals and conferences such as Nat. Mach. Intell., Nat. Comput. Sci., Nat. Commun., Nat. Biomed. Eng, NeurIPS, and ICLR, with citations exceeding 4800. Several of his achievements have been reported by renowned media outlets including MIT Tech Review, Forbes, China Science Daily, People's Daily and Xinhua Net. He has been selected for the Asian Young Scientist Fellowship, Forbes 30 Under 30 Asia, the WAIC Yunfan Award, and the Shanghai Chenguang Program. He has also received honors and awards such as the World Artificial Intelligence Conference Outstanding Paper Award, Rey Wu Prize, Baidu Scholarship, and CAAI Outstanding Doctoral Dissertation Award.




